This idea has been in my head for some time. I think RomanM deserves the credit for the concept, I have simply applied it to tree latewood density data. The idea is to use the actual “thermal hammer” least squares functions to provide a minimized sum of squares of errors fit to tree data. For readers who don’t recall the method, I have several posts on it:
http://statpad.wordpress.com/2010/02/19/combining-stations-plan-b/
http://statpad.wordpress.com/2010/02/25/comparing-single-and-monthly-offsets/
http://statpad.wordpress.com/2010/03/04/weighted-seasonal-offsets/
http://statpad.wordpress.com/2010/03/18/anomaly-regression-%E2%80%93-do-it-right/
https://noconsensus.wordpress.com/2010/03/21/demonstration-of-improved-anomaly-calculation/
https://noconsensus.wordpress.com/2010/03/24/thermal-hammer/
https://noconsensus.wordpress.com/2010/03/25/thermal-hammer-part-deux/
Since it is unlikely that many will read those links, the idea behind them was to take care of the anomaly calculation and the offset of temperature station data in different altitudes and regions with a single step least-squares calculation. So when looking for a global average, a colder mountain based station can be combined with a valley station in a highly optimized manner.
Well, some years ago, Roman left a comment about applying the methods to proxy data. I found the comment interesting enough that it hasn’t disappeared from my thoughts over all of that time. One of the main concerns in dendroclimatology is finding the most optimal method for combining series of tree data such that long term signals are not repressed.
Example:
Temperature is rising from 0-1 C over 100 years.
You have two thermometer trees (trees supposedly responding to temperature linearly with their growth) and they measure such that the first tree grows from year 0-55 and the second tree from year 45-100 so each tree has an overalap period from years 45-55.
Results of different methods:
First, the actual temperature which occurred and the two perfect trees which recorded it. Between years 45 and 55 the black line is covered by the red line. 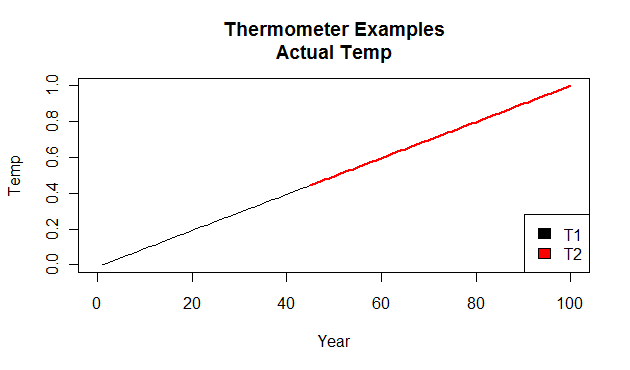
Now the typical assumption is that each nearby tree may respond a little differently to temperature due to factors such as soil condition or local competition. This makes the calibration of each tree a separate calculation. In my previous post, I used a mean of the tree ring size and got a very flat looking reconstruction with no substantial deviations from an average growth rate. The calculations subtracted the mean of each tree and scaled them by standard deviation before averaging. Subtracting the mean is a standard practice in the field of dendrochronology prior to a variety of ad-hoc methods to restore the long term variance. If we don’t work to restore long-term variance, the following two plots show what happens.
First there are two overlapping thermometer trees with means subtracted.
 This is what the mean looks like after centering:
This is what the mean looks like after centering:
 Of note is that while the series is now centered, the original temperature experienced by these thermometers was 0 to 1 degrees, and this curve is from -0.27 to 0.27. We can see that the long term variance (signal) is suppressed by the method.
Of note is that while the series is now centered, the original temperature experienced by these thermometers was 0 to 1 degrees, and this curve is from -0.27 to 0.27. We can see that the long term variance (signal) is suppressed by the method.
Using the least-squares offset calculation functions introduced in the thermal hammer method, we can take the two offset treemometer series and calculate the ideal offset for each series. The result is shown in the next graph:

The data is from -.5 to 0.5 or exactly 1 and has no steps in the center where the data from the two series overlaps. The great bit about this multi-variate least squares approach, is that you can literally ‘stuff’ as many series as you want into it and find the minimum least squares error for every single one.
—–
Growth curve:
The meat of the post in this case is very long and takes some explanation. I’m going to brush by a few concepts without a lot of explanation, those with familiarity in dendro issues won’t have trouble with this but others may.
First, I piled the polar.mxd latewood density data into a matrix by tree age and calculated a growth curve by mean of the year. This curve has annual level noise which is clearly not related to the age of the tree (growth curve). I calculated the curve different ways but personally it is an ad-hoc correction for something we know exists so my favorite correction is the simple way of just low-pass filtering the signal. Spline or other fits make little difference. For this data, we did demonstrate previously that there is a signal in the growth curve that should to be corrected for, still some may not like the correction I used as it can cause unwanted effects, so I performed the following calculations both with and without age-based corrections.
Method:
I provide software below so readers can check whether my methods are what I claim.
Trees (123 core samples) are arranged into a timeseries of 400 x 123 with each tree starting at year zero
Age curve is calculated by averaging the tree density value for each year
Filtered age curve is subtracted from each tree in no growth signal matrix – ngsalltrees
Trees are compiled into larger matrix with start year being actual year in which tree grew. Other values are NA.
No offset simple reconstructions were done by taking row means.
Least squares reconstructions were then done to minimize least squares error overlapping data for entire matrix.
Results:
First, we have the simple mean reconstructions without and with growth curve corrections. Note the flatness of the long term signal.
The next graph includes the age based growth curve correction.
Then we start to get to the important part of this post. The first least-squares reconstruction from tree data that I have seen anywhere. To be clear, this is the same data which produced the previous two graphs.
This next plot has the growth signal removed from each tree, is offset by least squares. Conclusions:
Conclusions:
I think there is plenty to discuss about the shape of this curve and whether it has anything to do with temperature at all. From our previous posts, we know there is a good quality high frequency match to regional temperature data. I will need to take some time to see if trends match better. While I am still highly skeptical that this is a representation of long term temperature, this method is far more objective than the ad-hoc iterative approach used in the Briffa 2013 “no-signal” dendrocrhonolgy paper.
While I’ve noticed the propensity for people to look at these graphs and assume they are temperature, I must warn that the limitations of this method for recreating long term trends are significant. Each series knits with residual error to the other series so that as we go further from the calibration period in time a “walk” in the series occurs. Since calibration is in recent years when thermometers exist, growth curve accuracy is lost in history. I think some sensitivity analysis of the method to added noise and different growth curve strategies is in order. If other changes don’t generate significant differences though, and I don’t think they will, the last curve is simply the data and either we accept that it is temperature only or something else.
Each method used in proxy history seems to have its own disadvantages, no-signal dendrochronlogy of Briffa 2013 represses the effect by removing the average signal in as the growth data and employing an average. While the no-signal method will regularly produce a less dramatic looking chronology, it cannot capture long term variance from tree growth in significantly different altitude or soil conditions.
Although this is the first time I have ever seen this method used in proxy based reconstructions, I cannot take credit for it as RomanM of Climate Audit fame came up with the foundational concept.
—
I will post the code later tonight!
#ROMAN M LEAST SQUARES OFFSET FUNCTIONS
psx.inv = function(mat,tol = NULL)
{
if (NCOL(mat)==1) return( mat /sum(mat^2))
msvd = svd(mat)
dind = msvd$d
if (is.null(tol))
{
tol = max(NROW(mat),NCOL(mat))*max(dind)*.Machine$double.eps
}
dind[dind
dind[dind="">0] = 1/dind[dind>0]
inv = msvd$v %*% diag(dind, length(dind)) %*% t(msvd$u)
inv
}
### subfunction to do offsets
calcx.offset = function(tdat,wts)
{
## new version
nr = length(wts)
delt.mat = !is.na(tdat)
delt.vec = rowSums(delt.mat)
row.miss= (delt.vec ==0)
delt2 = delt.mat/(delt.vec+row.miss)
co.mat = diag(colSums(delt.mat)) - (t(delt.mat)%*% delt2)
co.vec = colSums(delt.mat*tdat,na.rm=T) - colSums(rowSums(delt.mat*tdat,na.rm=T)*delt2)
co.mat[nr,] = wts
co.vec[nr]=0
psx.inv(co.mat)%*%co.vec
}
#### load external functions filtering used
source("http://www.climateaudit.info/scripts/utilities.txt") #Steve McIntyre
### Gausssian filter
ff=function(x)
{
filter.combine.pad(x,truncated.gauss.weights(51) )[,2]#31
}
### load briffa data
loc="c:/agw/briffa 2013/data/raw/polar/polar.mxd"
wd=c(12,6,6,6,6,6,6,6,6,6,6)
dat=read.fwf(loc,widths=wd)
treename=substr(dat[,1],1,8)
treeid=levels(factor(treename))
year = as.numeric(substr(dat[,1],9,12))
allyear=levels(factor(year))
alltrees=array(NA,dim=c(400,length(treeid)))
treemin=rep(NA,length(treeid))
### align trees by age rather than year
for (i in 1:length(treeid))
{
mask= treename==treeid[i]
da=dat[mask,]
yr=year[mask]
treemin[i]=min(yr)
for(j in 1:length(yr))
{
ageindex=yr[j]-treemin[i]+1
alltrees[ageindex:(ageindex+9),i]=as.numeric(da[j,2:11])
}
}
mask= alltrees== -9999 | alltrees== -9990
alltrees[mask]=NA
alltrees=ts(alltrees,start=1)
#plot(alltrees[,1:10])
### center and normalize all trees by standard deviation
alltrees=t(t(alltrees)-colMeans(alltrees,na.rm=TRUE)) #center
alltrees=t(t(alltrees)/sd(alltrees,na.rm=TRUE)) #normalize to sdev
alltrees=ts(alltrees,start=1)
## calculate growth curve
## this curve is an age based mean value of each tree's growth
## The curve is low-pass filtered to remove high frequency non-age related noise
par(bg="gray90")
growthcurve=ff(ts(rowMeans(alltrees,na.rm=TRUE),start=1))
plot(growthcurve,main="Growth Signal from Briffa 2013 Polar MXD Data",xlab="Age (years)",ylab="Unitless",ylim=c(-1,1))
##no growth signal version of alltrees
ngsalltrees=alltrees-growthcurve
## create year based matrix from no growth trees start year is 870
tsdat= ts(matrix(nrow=1800, ncol=length(treeid)),start=870)
ngtsdat=ts(matrix(nrow=1800, ncol=length(treeid)),start=870)
for(i in 1:dim(ngsalltrees)[2])
{
yearindex=treemin[i]-870
print(yearindex)
tsdat[yearindex:(yearindex+399),i]=ngsalltrees[,i]
ngtsdat[yearindex:(yearindex+399),i]=alltrees[,i]
}
#################
# no growth calculations
ngaveragereconstruction=window(ts(rowMeans(ngtsdat,na.rm=TRUE),start=870),end=2010)
averagereconstruction=window(ts(rowMeans(tsdat,na.rm=TRUE),start=870),end=2010)
plot(ngaveragereconstruction,main="Simple Mean Reconstruction\nFiltered Growth Curve Correction",xlab="Year",ylab="Unitless Growth")
lines(ff(ngaveragereconstruction),col="red",lwd=3)
plot(averagereconstruction,main="Simple Mean Reconstruction\nNo Growth Curve Correction",xlab="Year",ylab="Unitless Growth")
lines(ff(averagereconstruction),col="red",lwd=3)
plot(ngaveragereconstruction)
plot(averagereconstruction)
# Least squares offset reconstruction
a=calcx.offset(tsdat,rep(1,dim(alltrees)[2])) #.1711838
tsdatoffset=t(t(tsdat)-(as.vector(a)))
lsreconstruction=window(ts(rowMeans(tsdatoffset,na.rm=TRUE),start=870),end=2010)
plot(lsreconstruction,main="Least Squares Offset Reconstruction\nNo Growth Curve Correction",xlab="Year",ylab="Unitless Growth")
lines(ff(lsreconstruction),col="red",lwd=3)
# Least squares offset reconstruction
a=calcx.offset(ngtsdat,rep(1,dim(alltrees)[2])) #.1711838
ngtsdatoffset=t(t(ngtsdat)-(as.vector(a)))
nglsreconstruction=window(ts(rowMeans(ngtsdatoffset,na.rm=TRUE),start=870),end=2010)
plot(nglsreconstruction,main="Least Squares Offset Reconstruction\nFiltered Growth Curve Correction",xlab="Year",ylab="Unitless Growth")
lines(ff(nglsreconstruction),col="red",lwd=3)




Jeff,
Might some global trend migrate into the growth curves?
I wondered what would happen if you allowed growth curves for your case of the two perfect treemometers?
In my previous posts, the growth curve from pre-1500 trees was visually the same as post 1500. I was a bit surprised by its consistency. I need to find another MXD dataset and see if the result is similar.
What do you think about the concept of ls fit to knit the proxies?
Jeff, I think it is a good idea, in principle. The query is, how does the LS discriminate the components of the linear model. With Roman’s method, there’s a constant that you can’t determine that could be part of the offsets, or part of the signal. You have to add something to determine that.
Here it seems to me that “offset” is more complicated, since it could include the growth curve. I’m not sure if that creates more dof that could be part of the fitted “offset” or part of the signal.
Nick,
Thanks for the reply. I don’t think I’m following what you are saying though.
We are looking for a multi-lifespan signal in the tree ring history. We know that this signal is comprised of all kinds of environmental variables but hope that these trees are mostly temperature limited. We also know that individual trees have an age related growth rate signal that we would like to correct for. There is no math which can reliably separate any of the signals which are on the multi-century timeframe but signals related to tree growth can be detected separately from the multi-century signal by aligning the samples by setting the first tree ring to year 1 and averaging.
The offsets in this method align the samples to the best fit of the data and I found similar reconstruction trends using both growth corrected and uncorrected data. I don’t expect growth curves to alter the result much because there is enough overlap of the series.
Now you asked before if the growth curve would have long term signal in it. Of course you are right, some component of the signal is in it. However, by aligning the data and averaging rather than using a spline, I think we have removed most of the artificial stiffness that an equation fit would cause and have nearly maximized our separation of growth signal and multi-century trend. So when you ask for “offsets” vs “signal”, I’m having difficulty figuring out what you are suggesting.
You did make the point that offset should include the growth curve. I think we have already maximized that relationship in the last graph and unless we incorporate a curve fit growth equation as a function of tree age into the least squares fit, we cannot match the result. If that is what you meant, I would say that the insertion of that function is mathematically equivalent to the subtraction we have already done.
Nick:
Or perhaps the growth correction could include climate and therefore distort the climate offset calculations.
Interesting effort, Jeff. Glad to see you doing statistics again.
One minor typo in the R script: Line 11 should be replaced by the two lines:
dind[dind0] = 1/dind[dind>0]
Such errors happen to me all of the time when I start typing in what I believe to be the console window after running a portion of a script. I then waste time figuring out what damage I may have done to my script afterwards. 🙂
There are several possible approaches for dealing with the growth curve. For example, one can iterate alternately between estimation of the growth curve and the reconstruction until the situation converges. I have some questions in this case with how the data should be “standardized”. With the lower bound of 0 for all of the values, I am not sure that mean =0, sd =1 is the way to go.
There is also the question of whether growth should be subtracted from or divided by the estimated growth curve. In the latter case, this would suggest that using analyzing the logs of the data logarithms could be a reasonable approach as part of the overall analysis.
Lots of avenues to look at, but that is the fun on the whole venture.
Oops: TWO lines not one:
dind[dind0] = 1/dind[dind>0]
Sorry!
Screwed by the system again! It’s those “less than” signs interfering with the HTML. Try something different this time:
dind[dind0] = 1/dind[dind>0]
Thanks, I got the line fixed.
“one can iterate alternately between estimation of the growth curve and the reconstruction until the situation converges.”
This is the ‘no signal’ method of Briffa 2013. From reading the dissertation of a coauthor who’s name I can’t recall at the moment, it was quite the big advancement a few years ago.
Amazing! I am dumb as a brick when it comes to “R”.
Looks to me like a multi-dimensional optimisation problem, something I have been doing at various times over the past 30 years (a lot for real in commerce, it works, get paid). I concluded that most of the time on hard problems a crude method works, forget fancy maths, put the effort into high speed code. R will have libs, no idea on goodness.
Fundamentally, can you write a merit function? If you can wrap it in an optimiser loop which twiddles things. I assume that is what you have done.
If I guess correctly simply adjusting offsets is easy and you do it. The question seems to be whether you can adjust offset and the shape of each tree. I doubt it simplistically and this is heuristics. I would expect the growth to change with conditions (and possibly genetic selection, an involved subject given “mother” trees). This suggests some kind of local optimisation, not first tree against last if conditions are different… which depends on the final answer. Ouch.
This looks like an ambiguous problem which will have various sensible solutions. Experiment. beauty contest time.
But what do I know, barking mad up a tree.
Finally really off the wall. An oddity I have come across is where least squares fails. I first came across this with a manufactured test dataset, as recall, I hope, comprised several sines and a linear trend, now hammer it with random data deletions making a missing data point test. So severe it is difficult to recognise even if you know the answer. This was swallowed by discovery software. The oddity as I recall was least squares produced a stepped result, not a linear trend, a bizarre effect. Switching to a fourier merit function produced a straight line. This was bemusing, tie knot in brain, curiosity.
Quite recently I discovered a similar effect between 19th centry maths as used in meteorology etc. and using least squares… so I switched to fourier, curiously different result without what looked like artefacts, quantisation.
What is going on? Okay… how should I know? Just seems apt to mention it.
No optimizer loop. Just a multi-variate LS fit to 123 very noisy timeseries which cover different timeperiods. I completely agree with your comment on simplicity which I why I like this result over the standard in the field. I am not yet convinced the resulting timeseries means much of anything. Further study is required but I think it may be a good representation of the data. If it is a good representation of the data, I think it might be very strong evidence that this data is not responding primarily to temperature – as has been premised.